6 de October de 2024
Introduction to sabinaNSDM: A new R package to improve species distribution models based on spatially nested hierarchical models
Teresa Goicolea, Alejandra Zarzo
Species Distribution Models (SDMs) are essential tools for scientists and conservationists to predict where species are likely to be found, where they have existed in the past, and where they may appear in the future. With pressing issues like climate change and biodiversity loss, generating accurate predictions is more important than ever to identify key areas for conservation actions. However, SDMs often face accuracy issues, especially due to niche truncation and environmental extrapolation problems.
This is where the new R package sabinaNSDM comes in. Designed by our research team SABINA, this package uses a new approach to building SDMs, known as spatially nested hierarchical models (N-SDMs). By combining large-scale global patterns with finer regional features, sabinaNSDM allows for more accurate predictions of species distributions. This makes the new package a powerful resource for conservation planning and ecological research.
The problem with traditional SDMs
Standard SDMs present a set of limitations. Most models fall into one of two categories: regional or global.
- Regional models focus on specific areas, such as a country or a region. While they can offer detailed information about local conditions, they lack the broader environmental perspective that shapes a species’ distribution. This leads to what is known as niche truncation, where models do not consider the full range of conditions a species experiences across its distribution (i.e., its ecological niche). These spatially restricted models also suffer from a higher proportion of non-analogous conditions, leading to issues when projecting to other areas (e.g., predicting the spread of invasive species) or time periods (e.g., forecasting the impact of climate change on species distribution).
- On the other hand, global models cover an entire species’ range but often rely on coarse and low-resolution data. They also typically rely only on bioclimatic variables, as other environmental factors are unavailable at such a large scale, and the species data are often imprecise. As a result, they lack the fine details needed for accurate localized predictions.
The solution: Nested Species Distribution Models (N-SDMs)
Spatially nested hierarchical SDMs (N-SDMs) address these issues by combining the broad perspective of global models with the fine detail of regional models to get the best of both worlds. Global models provide an overview, capturing a species’ full ecological niche across its range, and take into account factors like climate at a coarse resolution. Regional models then focus on finer details, such as land use or microhabitat conditions, and more precise species distribution data, which are usually available for smaller areas, such as at the national level. These fine details are critical for making accurate and high-resolution predictions.
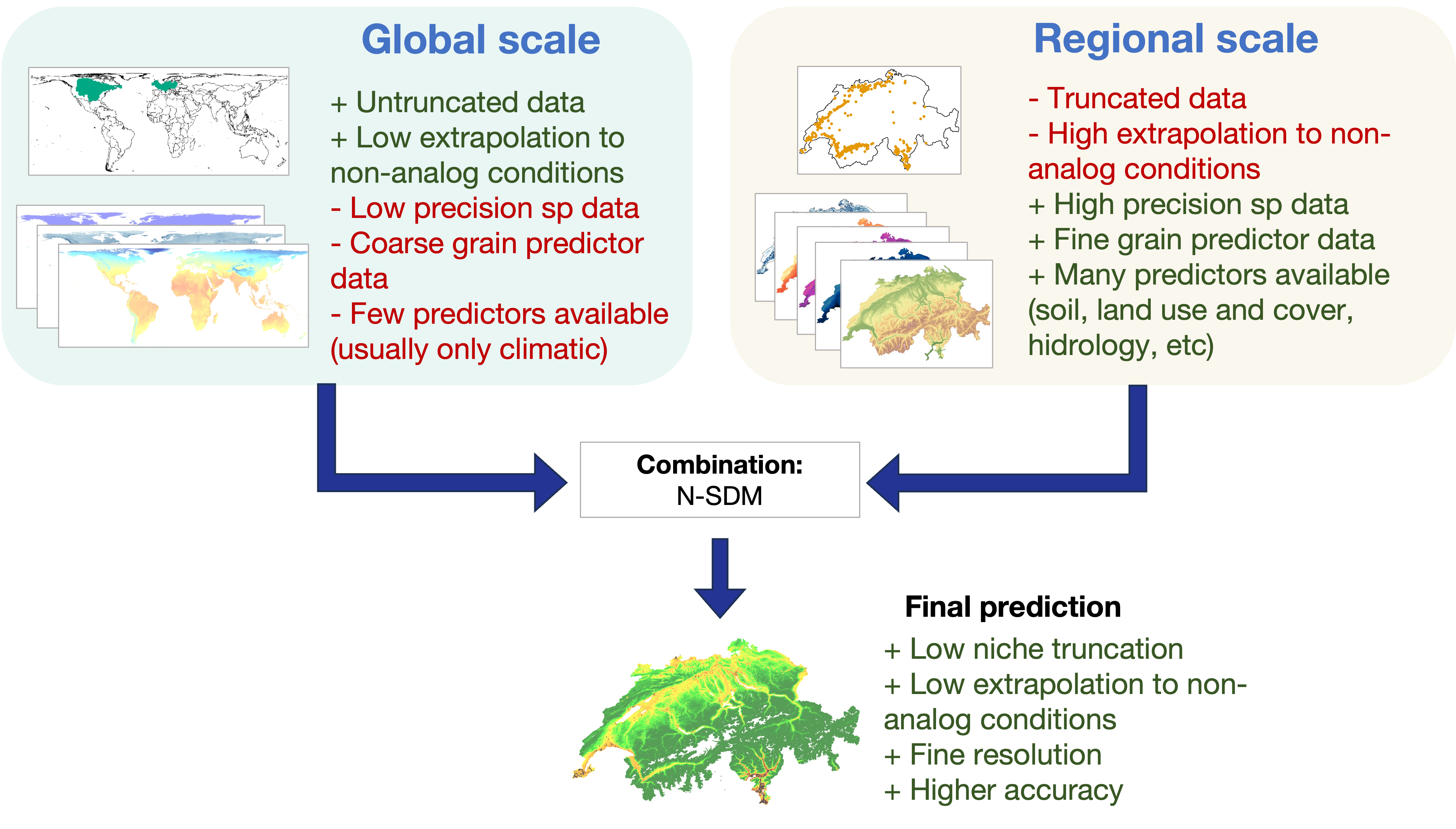
Figure. Advantages (in green) and limitations (in red) of traditional species distribution models (both global and regional scales), compared to the benefits of combining them into a Spatially Nested Species Distribution Model (N-SDM).
Key features of the sabinaNSDM package
sabinaNSDM is designed to make the N-SDM approach more accessible to researchers and conservationists. Here are some of its key features:
- Generate N-SDMs: The package combines global and regional models.
- Different nesting strategies: Users can choose between two methods for combining models: the covariate approach, which uses the output of the global model as input for the regional model, or the multiple approach, which averages the global and regional predictions.
- Consensus models: sabinaNSDM uses consensus models, a technique that combines multiple statistical algorithms to increase prediction reliability and accuracy.
- Comprehensive workflow: The package is a tool that integrates (a) background data generation; (b) preparation and spatial filtering of species occurrences (and absences, if available); (c) environmental covariate selection; and (d) N-SDM calibration, evaluation, and projection.
- Proven effectiveness: In an applied study on 77 tree and shrub species in the Iberian Peninsula, sabinaNSDM outperformed traditional SDMs, providing more accurate predictions of these species’ distributions.
- Open-source and user-friendly: sabinaNSDM is freely available on GitHub, and we are working to make it available on CRAN. The package is designed to be user-friendly, making it accessible to ecologists and conservationists with varying levels of programming experience.
Real-world impact
The ability to accurately model species distributions has real-world consequences, and the improved modeling capabilities of sabinaNSDM can play a crucial role in guiding conservation efforts more effectively. For example, the package can predict how climate change may alter species distributions, guide restoration programs to identify areas with the greatest potential to protect biodiversity, or anticipate the spread of invasive species. One of our key applications has been creating a geoportal that shows the predicted distribution of 200 woody plant species in Spain under current conditions and four future climate scenarios. The geoportal offers practical applications, such as generating lists of shrubs and trees with the highest suitability for specific locations. This can help inform restoration efforts by identifying species most likely to thrive both now and in the future. sabinaNSDM has already demonstrated its potential in our work, and we are excited to see how other researchers and conservationists use it in their projects.
Start using sabinaNSDM
If you are interested in trying sabinaNSDM, you can download the package and explore its features in our GitHub repository. For a deeper dive into how it works, check out our article published in Methods in Ecology and Evolution. We have also included supplementary material and tutorials to help you get started with single or multi-species models. If you are interested in learning more about sabinaNSDM or have any questions, feel free to get in touch.




